Nanoelectronics for DNA Sequencing
DeoxyriboNucleic Acid (DNA) is a molecule which carries the inheritable genetic information, a blueprint for each living organism. Double stranded DNA is a combination of two antiparallel polymers, strands, built of mutually weakly bonded monomers, DNA nucleotides, each made of a sugar-phosphate backbones to which are attached DNA bases (in figure below). There are only four types of the base molecules, adenine (A), guanine (G), cytosine (C) and thymine (T).
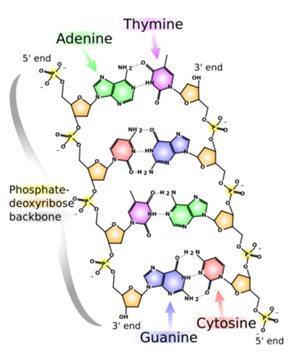
Figure 1: A segment of SSDNA
The sequence of the nucleotides in a segment of DNA, gene, caries the genetic information. A human DNA, human genome, may contain a few billions bases but only tens of thousands genes. The pairing of DNA nuclotides between the two antiparallel polymers, which creates its well known double stranded helical structure, follows particular rules, for example A pairs to T, G to C, etc. Therefore, DNA sequencing consists of determining the order of bases on a single strand, for example, ACTG... in the left strand (figure 1, counting from the top).
DNA sequencing is one of the leading precursors of the personalized medicine, i.e. use of individual genetic code to prevent, diagnose and treat diseases. Besides medical diagnostic, it has a large importance in biotechnology and in forensic sciences. The next generation of genome sequencing technology is expected to bring the cost (<$1000) and time (< 24h) for sequencing a human genome into a range that is affordable for diagnosis and prognosis related to individual patients.
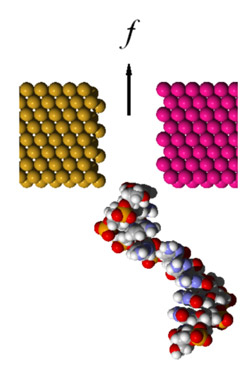
Figure 2: A DNA translocation through a nanogap
The development of the DNA sequencing technology is a collective effort which integrates biology, chemistry, physics and engineering. Particularly interesting is the prospective of physics-based methods, since they are intrinsically fast and can operate on a single DNA molecule. Basic idea of the physical methods 1) is electrical detection of the DNA sequences by reading the transversal electron tunneling current between the electrodes while a DNA is translocated through a synthetic nanopore or a nanogap by an electrophoretic force (schematically, as in figure 2). The measurement of the conductance of a single molecule is an attractive concept for molecular detection because single-molecule conductance may be governed by its intrinsic electronic properties. The main challenges in this approach are twofold, namely, 1) electric signal-to-noise ratio that distinguishes between the DNA bases, and 2) full control and localization of a DNA while it is translocated through the nano-electrode gap or nanopore.
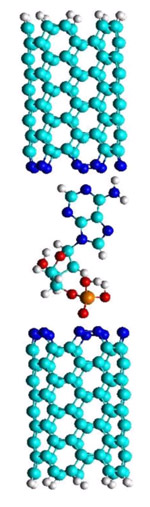
Figure 3: N- doped CNT as a DNA probe
Charge transport, in this case electron transport, through biological molecules ultimately depends on the electronic and chemical structure of the molecules, altered by the presence of the electrodes. The electronic and chemical structure of the four DNA nucleotides (A, C, G, T) are intrinsically different, and should, in principle, produce distinguishable electron transport properties while DNA is translocated through a nanogap between nanoelectrodes, as shown above. Assuming existence of a practical way for stable threading a single stand (ss) DNA polymer through the nm gap, the DNA sequence could be read from conductivity measurements as a function of the position of the polymer in the gap. Classical molecular dynamics simulations have shown that it is possible to "pull" a DNA through a nano-electrode gap by applying the electrophoretic force along the gap.
However, ssDNA experiences significant deformation and stretching when it is pulled through the gap. Furthermore, an across-the-gap E-field of 100mV/nm is enough to attract and "glue" DNA to a panel electrode surface and prevent control of the DNA translocation. In addition to these difficulties, the thermal fluctuations, Brownian motion, transversal and electrophoretic fields, cause strong fluctuations of instantaneous mutual position of the DNA bases and the electrodes, i.e. a large noise and poor signal-to-noise ratio in the transversal nonresonant tunneling conductance. Concerning improvements of the DNA translocation control, there are several approaches such as translocation through a Paul nanotrap or through a narrow CNT, in aqueous/electrolyte environment.
One approach to increase the sensitivity of the sequencing and signal-to-noise ratio is to functionalize the carbon-nanotube (CNT) electrodes, i.e. to dope them selectively (figure3). This fictionalization results in a signal enhancement of several orders of magnitude and is a consequence of resonant tunneling created by increased coupling of the doped nitrogen by the HN and HO groups in a nucleotide (H-bond). There are also other ideas of functionalization, all based on the relatively weak H-bonding, which shifts the levels of the expanded molecule at figure 3, toward the Fermi energy of the electrons in the electrodes, providing quasiresonant conditions for the electron tunneling (figure 4). The "red" energy levels are centered at the nucleotide thus enabling a quasi-resonant "hoping" of electrons from the electrode to the nucleotide, ending at another electrode. As can be seen in figure 4, functionalization with hydrogen (in place of nytrogen) does not create quasiresonant conditions. Detecting single chemical and biological molecules without using receptors or chemical interfaces is a paradigm shift in chemical sensing. This approach may also pave the way for developing receptor-free sensing concepts that are purely electrical.
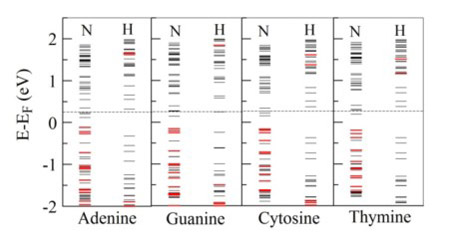
Figure 4: Molecular energy diagram of the probe at figure 3
In 2006, the X Prize Foundation, established the Archon X Prize for Genomics which will award $10 million to "the first Team that can build a device and use it to sequence 100 human genomes within 10 days or less, with an accuracy of no more than one error in every 100,000 bases sequenced, with sequences accurately covering at least 98% of the genome, and at a recurring cost of no more than $10,000 per genome. [1]
Acknowledgement

The content of the web page has been created by Dr. Predrag Krstic from Oak Rudge National Lab.
References
[1] D. Branton et al, "The Potential and Challenges of Nanopore Sequencing," Nature Biotechnol. 26, 1146 (2008)
More about DNA Sequencing
- DNA Sequencing - Wikipedia Article
- DNA Sequencing - by John Kimball
- How Do We Sequence DNA?
- Essential Biochemistry - DNA Sequencing
- Single Molecule Real Time DNA Sequencing (Video)
- Facts About Genome Sequencing
- A Brief Guide to DNA Sequencing
